The recent proliferation of digital microfluidic (DMF) biochips has enabled rapid on-chip implementation of many biochemical laboratory assays or protocols. Sample preprocessing, which includes dilution and mixing of reagents, plays an important role in the preparation of assays. The automation of sample preparation on a digital microfluidic platform often mandates the execution of a mixing algorithm, which determines a sequence of droplet mix-split steps (usually represented as a mixing tree/graph).
In general, for (m:n) mixing model used in each mix-split step on a DMF biochip, if C_h and C_\ell are two concentration factors (CFs) of two input fluids, then the CF of the resultant droplets can be written as
In case of dilution of a fluid, we are supplied with ONLY TWO input fluids and we consider the second fluid as the buffer solution. Whereas, mixing is a general term, in which we can mix TWO or MORE different reagent (input) fluids together step-by-step to achieve or approximate a target ratio of CFs of input fluids.
If the target ratio (volumetric) of N different reagent fluids is given as x_1:x_2:\ldots:x_N = a_1:a_2:\ldots:a_N, where the variable x_i denotes reagent fluid and a_i denotes the coefficient corresponding to the variable x_i in target ratio, then an algebraic expression A(N) can be written as follows to represent the CFs of N reagent fluids in the target mixture:
Dilution of a sample fluid is one of the basic steps required in almost all bioprotocols. However, in case of DMF biochips, as the biochemical fluids can only be mixed using discrete volumes of droplets, it is possible to carry out only serial dilution to achieve or approximate desired non-linear CFs. Note that the dilution process needs only two input fluids (sample and buffer) to prepare a target solution, and up to two mixers are sufficient to complete the task. A number of algorithms have appeared in the literature for performing automated dilution of a sample fluid (CF = 100%) to a certain CF by mixing it with a buffer (CF = 0%) on a DMF platform as follows: [Ren et al., ISSAMC, 2003], DC [Griffith et al., TCAD, 2006], twoWayMix [Thies et al., Nat. Comp., 2008], DMRW [Roy et al., TCAD, 2010], IDMA [Roy et al., DATE, 2011], MD [Bhattacharjee et al., ISED, 2012], REMIA [Huang et al., ICCAD, 2012], GORMA [Chiang et al., VLSIDAT, 2013], WARA [Huang et al., TCAD, 2013], NFSP [Dinh et al., ASPDAC, 2014], MTC [Mitra et al., TCAD, 2014], IDS [Roy et al., Integration, 2015].
Mixing Algorithms:A number of algorithms have appeared in the literature for performing automated mixing of two or more reagent (input) fluids on a DMF platform such as MinMix [Thies et al., Nat. Comp., 2008], RMA [Roy et al., VLSID, 2011] [Roy et al., TODAES, 2015], RSM [Hsieh et al., TCAD, 2012], MTCS [Kumar et al., DDECS, 2013], CoDOS [Liu et al., ICCAD, 2013].
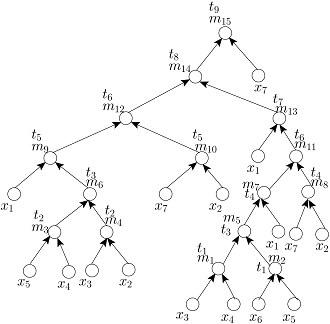
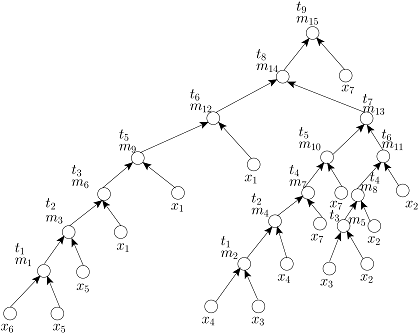
Given the target ratio as 20:10:2:2:2:1:53 of 7 reagent (input) fluids x_1,x_2,x_3,x_4,x_5,x_6,x_7 for an example of automated mixture preparation using a DMF biochip. FOUR mixing trees determined by running FOUR mixing algorithms such as MinMix [1], RMA [2], MTCS [4], and CoDOS [3] are shown below:
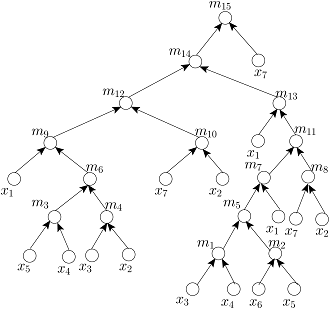
MinMix Tree
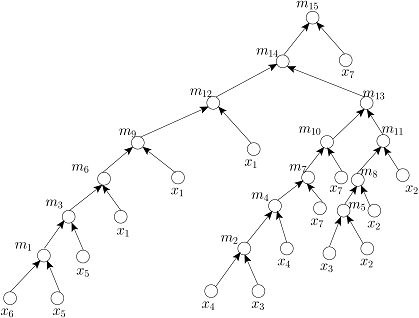
RMA Tree
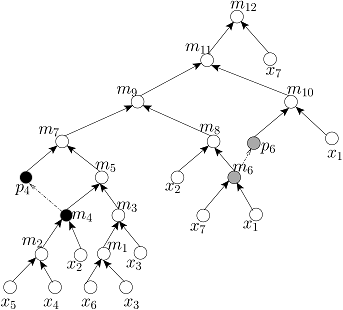
MTCS Tree
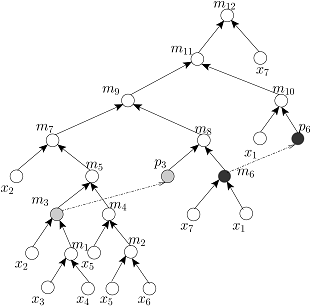
CoDOS Tree
Synthesis of a DMF biochip involves two steps: architectural-level (high-level) synthesis and geometrical-level synthesis (physical design) [Chakrabarty et al., CRC Press, 2007]. Architectural-level synthesis of DMF biochips involves bioassay scheduling, resource binding and sharing. the optimization problem of bioassay scheduling under resource constraints is formulated, which determines the start and stop times of all the assay operations, subject to the precedence constraints imposed by the sequencing graph. However, since this scheduling problem is NP-complete, heuristic techniques were developed to solve the problem in a computationally efficient manner. In a valid schedule, assay operations that share a resource cannot execute concurrently. Due to resource constraints, a resource binding may associate one functional resource with several assay operations of the same type; this necessitates resource sharing [Chakrabarty et al., CRC Press, 2007].
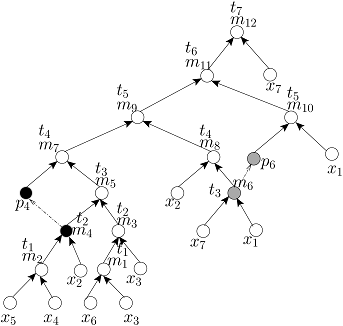
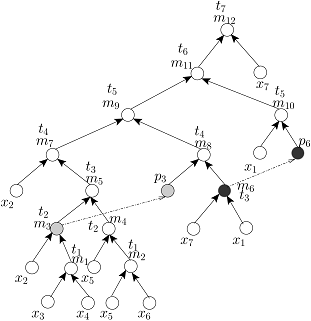
Without any loss of generality, we assume that all (1:1) mix-split operations are identical and each of them can be executed by an on-chip mixer in unit time step. In order to schedule a given mixing tree on a DMF biochip, any existing algorithm, such as list-scheduling [Su et al., JETC, 2008], genetic algorithm based scheduling [Ricketts et al., DATE, 2006], path scheduling [Grissom et al., DAC, 2012], force-directed list scheduling [O’Neal et al., VLSI-SoC, 2012], or optimal scheduling with M mixers (OSM) [Luo et al., TASE, 2011] can be used. However, if the time-optimal scheduling algorithm namely OSM with M=2 (i.e., with TWO on-chip mixers) is used to schedule the above FOUR mixing trees, then the scheduled mixing trees will be as follows:
Scheduled MinMix Tree
Scheduled RMA Tree
Scheduled MTCS Tree
Scheduled CoDOS Tree
Given the ratio-sum (L), we have generated benchmark ratios (by varying the number of reagents from 3 to 12) using a number partitioning technique as described in [2]. Keeping only unique ratios (e.g., 1:1:2 and 1:2:1 are not unique), THREE files containing benchmark ratios for L = 16, 32 and 64 are available here for further use by anyone working in this domain:
Some Ratios used in Real-life Bioprotocols:
Here we share a FILE containing some ratios used in real-life bioprotocols along with with citations.
References:
